Overfitting Thriller!
So goofy I just had to share this “Machine Learning A Cappella – Overfitting Thriller!”
So goofy I just had to share this “Machine Learning A Cappella – Overfitting Thriller!”
(email to my fellow researchers)
This morning I completed the final evolutionary process for my GP code. It now supports Reproduction (no mutation), Point Mutation, Branch Mutation, and Crossover Reproduction.
I have run thousands of trees through the system without a glitch. In my new “debug” mode I am able to visually monitor the expansion and pruning of GP trees to make certain they are evolving as required.
I have learned the Sympy library which takes my full strings and reworks them to functional, more compact functions is not terribly forgiving. Yes, it will take a few dozen elements in a long polynomial and reduce the complexity (removing two instances of a variable which cancel each other, or rewording multiple instances as 2x, 3x, etc.), but if there is an error it simply stops execution.
This works to my favour, as I am able to trust the expressions are solid and fully functional in each generation.
I am a bit OCD so I will surely spend time making certain the internal documentation is clean and well stated. This will become the foundation for my User Guide and appendix in my Thesis.
The next major step is to run a few benchmark problems through my code to make certain I am seeing the desired output. Then, move into working again with the KAT7 data.
I am excited to get back to literature review, my thesis, and not programming every day :)
Thanks for all the support this past 3 months!
kai
I have been coding for nearly three months, with but a ten days break. I love this time and space. Singularly focused. Only one task in life. I leave my phone at home, launch my development Virtual Machine (no email, no Facebook), and dive in.
My machine learning (a subset of Artificial Intelligence) code “came to life” about ten days ago when it first converged on a solution. That was an incredible feeling. Almost there! Just one more type of mutation to complete. All tools are built but this one. Then it should just work.
While I set out to build a script to work with SKA KAT7 data, my tendency toward building things BIG kicked in and my GP instead became a platform. As a result of my approach, I inadvertently developed a pointer system for managing Numpy arrays. Each column in an axis-1 array is treated as a unique object with parent/child relations to others in the array.
If I had known how hard this would be, I would have looked for alternatives or considered a different plan. But sometimes it is best to just go into a battle blind, look back, and realize how far you have come.
When I arrived to Sutherland, with eight days and nights stretched out before me with just one principal task on my agenda, I was certain the Grow method (one of two branch mutations) was in my grasp. Yes, it evaded me for that week and another after I returned to Muizenberg. In fact, it became the single most challenging part of building Karoo GP (as noted in the email to my professor and fellow researchers May 26).
Despite the myriad sketches I produced, I was unable to see the path to fruition. I must have pursued a half dozen avenues, writing hundreds of lines of code that in the end, all failed.
I began to notice a pattern in my behaviour. I would repeatedly envision a solution. From that vantage point, I could see the challenge, the path, and the conclusion. I sketched the means, to assure it was real, and then dove into the code.
Several hours, even a day or two later, I hit a wall. Not just a small barrier, nor new challenge, but an insurmountable, unavoidable wall. With each wall, I was again reminded there was no work-around, no short-cut. The only solution invoked many stages and a great deal of coding.
How could I not have seen this? Why had this only now become no-go when just minute before, it was still possible?
I drew more sketches, more flow-diagrams, and arrived to more solutions. I even came back to an already failed solution. Having forgotten why it failed, I felt sure I must have missed something and dove back in only to be met with the same, obvious wall.
I felt stupid. Frustrated beyond belief. What’s more, my clarity of vision was diminishing rapidly. I could no longer see a half dozen steps down the coding path, but only two or three. My reduced exercise, pulling late nights with the astronomers, and change in diet all contributed.
I recall a high school art class in which we followed a book called “Drawing on the Right Side of the Brain by Betty Edwards” by Betty Edwards. One chapter advised us to hold a pencil or pen in our favoured hand, brainstorm and produce thumbnail sketches until our ideas ran dry. We felt we had surely exhausted all the possibilities for that concept.
Then we grasped the pen with the other hand, even if awkward to the majority of us who are not ambidextrous. More ideas came! Ideas from a completely different foundation.
I knew enough about my internal processes, having spent much of my undergraduate program in Industrial Design in problem solving, to recognise that I would benefit from working on something else, something that came to me more easily. I improved the interface, cleaned up the Python methods, and developed small, useful tools such as a method for tree population renumbering, another for renumbering columns in an axis-1 Numpy array, each of which represented a node in a GP tree; and ultimately, a method for tracking the differences between original and skewed node positions as a branch was inserted into a tree.
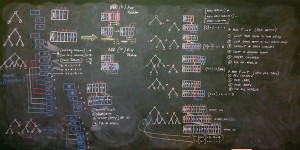 When I returned to Muizenberg and my office with a blackboard, I drew my diagrams much larger, using my entire upper body to bring form to my visions. I was able to see the solutions more clearly than when at Sutherland.
When I returned to Muizenberg and my office with a blackboard, I drew my diagrams much larger, using my entire upper body to bring form to my visions. I was able to see the solutions more clearly than when at Sutherland.
Yet, repeatedly, when I turned to sit at my computer, the blackboard to my back, the clarity vanished quickly. I’d start typing and … hit a roadblock, or completely forget what was to happen next.
I thought I was losing my mind. I could see two, sometimes three steps at a time, but four or more and I failed miserably. Endless loops of indigestible code, a headache, backache, and neck filled with anxiety.
I returned to the blackboard, erased everything, and started over. I took the sketches deeper, down to the pseudo-code level such that I was writing loose versions of Python directly on the board.
Those tiny tools I had developed in Sutherland became invaluable, for in each was a means by which I could relieve my mind of that function, trusting the power of a tested black box. The more tools I developed, the more I could focus on flow instead of the detailed interaction of ever level of code.
Success came to me when I broke a complex problem into the smallest pieces possible, connected those pieces with each other, and then validated the flow of data across the system. From this came the “debug” mode, not integral to Karoo GP as a means by which the user can monitor the mutations in real-time.
It’s fascinating to watch, even to me.
(email to my fellow researchers)
A week has come and gone. And with it, some 60 hours coding, if not more. I have accomplished a great deal, further fine-tuning the code, building tool-sets, and building a stronger method-based platform for GP. I can now build a Tree from the command line in 5 lines of code, evaluate for fitness, mutate, and save with a few more.
While I witnessed my GP converge on a solution a few nights ago for the first time, using only reproduction (copy) and simple point mutation, I have not yet completed crossover nor the grow method for branch mutation.
These have proved to be incredibly, mentally challenging. I have never struggled with something at this level (I want to say “in my life,” but I may not be thinking clearly :)
I freely admit the issue is not one of Python, but an incredibly complex array book keeping effort that borders on my having to create my own pointer system.
I spent 3 days exploring work-arounds, tricks, and short-cuts, but in the end, am back to the realisation there is just one way–the hard way.
The good news is that I have slowly eroded the challenge by building a suite of tools, each of which performs a single task. Cool.
I will get there, one step at a time.
Today was lost to the realisation (and subsequent rewrite) that my core Tournament code was (inadvertently) taking advantage of Python’s ability to perform basic comparisons (>, <, =) on strings which contains numbers, as compared to converting those to integers first. It works, but the results are not consistent. Fixed! I am far more confident, even though another day behind my slated sched.
What’s more, Python’s numpy random generator is clearly NOT random by any stretch of the imagination. 100 Trees evolved over 10 generations in non-interactive mode generates far, far more positive results than when I enter interactive mode (code pauses / continues with RETURN). This causes me to believe the random generator is using keyboard input as a random seed, which drastically (and negatively) affects the results.
I will, at another time, replace Numpy with a call to a proper, external random number generator. I see this current effort as computer science more than applied mathematics. However, what I am learning about the underlying structure of code is likely quite valuable.
Been coding till 2 or 3 am every night, photographing when the sky is clear ’till 5 or 6 am. Sleep till 10:30, go for a run, and back to the grind :)
kai
At roughly 1:45 am, my code came to life for the very first time. My genetic program found a solution not through a one-time, randomly generated polynomial expression, but through an evolutionary process which arrived to the desired solution.
I pushed back from the end of the SALT control room desk, threw my hands into the air, and at a respectable volume, “It’s alive!”
Given an equation with 2 terminals (variables) and 1 function (operand):
goal: a^b = c
data: 4,2,16
operands: +,-,*,/
Zero solutions found in the first 3 generations, but by the 5th, it produced 7 correct solutions out of 10 trees. This is with a very basic “either you get it or you don’t kernel”. No parsimony (reduction to the simplest solution). No crossover. Just simple point mutation. I also tried up to 100 trees in fewer generations or fewer trees in 100 generations. Each converges at a different rate.
I am very, very excited … and totally exhausted.

I was again mugged last week, just three train stops down from Muizenberg. My fellow researcher was knocked to the ground and repeatedly hit upside his head. I tried to defend, but one of the four guys displayed a gun tucked into the front of his trousers when I charged them. I backed off while one of them forced his hands into my friend’s pockets. Oddly, they did not discover his cell phone nor wallet. Following much yelling and insisting we had nothing for them to take other than the car battery we were carrying, they took the battery and ran.
We were lucky, we learned, as a man has been shot in that same spot a few weeks prior. Not the best part of town to walk through, it seems.
I was very much on edge for the following days. When I found a man who had been harassing another of the homeless population pressed up against a wall, the folds of his jacket in the clutches of my hands, I knew I had to get away for a while.
The South African Astronomical Observatory site at Sutherland is my retreat of choice.
Feels so good to be away from trains, sirens, car alarms, people shouting, and the constant awareness of potential crime. Planning a long hike across two or three ridges to a distant set of peaks I have wanted to visit since my first visit here two years ago.
Tonight I am working from the control room of SALT, one of the world’s largest telescopes. Listening to salsa, chatting with astronomers, and working on my Genetic Programming suite.
Every few hours I step out onto the wind-strewn plateau to enjoy the dark, southern sky.
(email to my fellow researchers)
Been coding 4-8 hrs every day for two weeks (since the GPU workshop). I feel I am making a snails-pace of progress, but at the same time learning a lot, step-by-step, piece-by-piece.
In the past 72 hrs I have migrated all functions into a Class (library), the foundation of proper Object Oriented Programming. Thuso stopped by yesterday and got me over a hurdle. Robert gave me some pointers as well. Mostly motivation. All coding remains at the tips of own fingers.
My code is clean, modular, and scalable. All functions are now ‘gp.[method]’ enabled. Once I get the internal message passing working properly, building a GP will be as simple as passing a few variables from one method to the next. In essence, I am creating a GP platform.
This may seem a bit overboard, but it sets the foundation for much simpler movement into the next steps: mutation, cross-over, and reproduction. I have merged what were two scripts into a single base which enables both Classify and Symbolic regression functionality, auto-selecting the associated dataset from the files/ directory.
The Tournament selection is done and tested. It randomly picks n Trees from the total population, selects the one with the highest fitness, and stores that in a list. Once for reproduction and mutation. Twice for cross-over.
Mutation is drafted. Need to call the methods in the right order.
Back to work …
kai