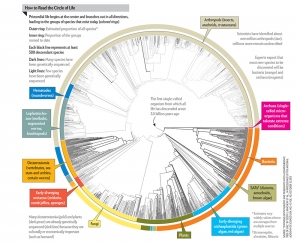
The Circle of Life
Toward the back of a recent issue of Scientific American, I was totally engrossed by a brief discussion of the “Circle of Life” from the point of view of biology. Every known species (2.3 million and growing) is included in the count (inner circle) with a projection for the balance of types of life, as we discover more, in the outer circle.
Perhaps what I love most is the understanding that we know so little, and are projecting our own lack of knowledge as a kind of map for what we desire, and will some day learn.